C11orf86
Chromosome 11 open reading frame 86, also known as C11orf86, is a protein-coding gene in humans.[1] It encodes for a protein known as uncharacterized protein C11orf86,[2] which is predicted to be a nuclear protein. The function of this protein is currently unknown.
Gene
Location
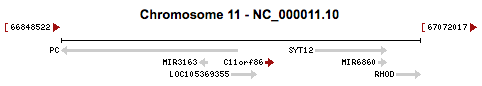
C11orf86 is located on the long arm of chromosome 11 at 11q13.2. It consists of 1732 base pairs, and is found on the plus strand. Gene neighbors of C11orf86 include uncharacterized LOC105369355, microRNA 6860, microRNA 3163, synaptotagmin 12, ras homolog family member D, and pyruvate carboxylase.[3]
Promoter
The program ElDorado, by Genomatix, identified the promoter region of C11orf86 on the positive strand from 66974707 to 66975464, for a total length of 758 base pairs.[4]
Expression
C11orf86 appears to be primarily expressed in the gastrointestinal tract.[5] Expression occurs in ascites, the intestine, the stomach, gastrointestinal tumors, and non-neoplasia.[6]
mRNA

Alternative Splicing
According to AceView, transcription of the gene produces three different mRNAs, two of which are alternatively spliced variants, while the third is an unspliced form.[7] All three variants could possibly code for functional proteins. The transcript used for this article is made up of two exons, amounting to 1185 base pairs, and has the reference number NM_001136485.1.[8]
| mRNA variant | Exon | Exon Size (bp) | Intron | Intron Size (bp) |
|---|---|---|---|---|
| a | 1 | 362 | 1 | 541 |
| 2 | 829 | |||
| b | 1 | 249 | 1 | 538 |
| 2 | 251 | |||
| c | 1 | 551 | 0 |
Protein
General Properties
C11orf86 protein is 115 amino acids in length.[2] The molecular weight of C11orf86 is 13.2 kdal.[9] Its isoelectric point is predicted to be 11.9.[10]
1 MGTGLRSQSL REPRPSYGKL QEPWGRPQEG QLRRALSLRQ GQEKSRSQGL ERGTEGPDAT 61 AQERVPGSLG DTEQLIQAQR RGSRWWLRRY QQVRRRWESF VAIFPSVTLS QPASP
Composition
The majority of the C11orf86 protein is composed of arginine (15.7%), glutamine (12.2%), serine (10.4%), glycine (10.4%), and leucine (9.6%). No cysteine, histidine, or asparagine residues are found in this protein.[9]
A : 6( 5.2%); C : 0( 0.0%); D : 2( 1.7%); E : 9( 7.8%); F : 2( 1.7%) G : 12(10.4%); H : 0( 0.0%); I : 2( 1.7%); K : 2( 1.7%); L : 11( 9.6%) M : 1( 0.9%); N : 0( 0.0%); P : 9( 7.8%); Q : 14(12.2%); R : 18(15.7%) S : 12(10.4%); T : 5( 4.3%); V : 4( 3.5%); W : 4( 3.5%); Y : 2( 1.7%)
C11orf86 has no positive, negative, or mixed charge clusters. However, there is a higher presence of arginine, which is positively charged.
1 00000+0000 +-0+0000+0 0-000+00-0 00++0000+0 00-+0+0000 -+00-00-00 61 00-+000000 -0-000000+ +00+000++0 000+++0-00 0000000000 00000
Domain
This protein is a part of the DUF4633 superfamily. Proteins that belong to this family are often between 94 and 123 amino acids in length.[11] This domain is found in bacteria, viruses, fungi, plants, insects, reptiles, birds, and mammals.[12]

Post-Translational Modification
C11orf86 is predicted to have nine possible phosphorylation sites, of which eight are serine, and one is threonine.[13] It is also predicted to have ten O-linked glycosylation sites.[14]
Secondary Structure
C11orf86 is primarily composed of random coil and alpha helices.[15][16]
Sub-cellular Localization
This protein is predicted to be a nuclear protein.[17] There appears to be a bipartite nuclear localization sequence beginning at position 80.[18]
Homology
The C11orf86 protein is conserved in mammals, and orthologs can easily be traced back to marsupials, monotremes, and reptiles. No orthologs of C11orf86 appear to be present in plants, fungi, fish, amphibians, or birds. There are no paralogs of C11orf86. The table below shows some orthologs that were found using BLAST.[19] Dates of divergence were found from TimeTree, using the median molecular time estimate.[20]
| Genus and Species | Common Name | Accession Number | Protein Length (aa) | Percent Identity (%) | Date of Divergence (mya) |
|---|---|---|---|---|---|
| Homo sapiens | Human | NP_001129957.1 | 115 | 100 | |
| Macaca mulatta | Rhesus macaque | XP_001115236.1 | 115 | 89 | 27.3 |
| Rattus norvegicus | Brown rat | NP_001102991.1 | 123 | 74 | 90.1 |
| Bos taurus | Cattle | NP_001070558.1 | 122 | 74 | 95.0 |
| Equus caballus | Horse | XP_014585069.1 | 113 | 71 | 95.0 |
| Mus musculus | Mouse | NP_081513.1 | 124 | 71 | 90.1 |
| Felis catus | Cat | XP_003993784.1 | 121 | 70 | 95.0 |
| Canis lupus familiaris | Dog | XP_003639729.1 | 121 | 69 | 95.0 |
| Sarcophilus harrisii | Tasmanian devil | XP_012408455.1 | 120 | 40 | 162.4 |
| Ornithorhynchus anatinus | Platypus | XP_007659213.1 | 119 | 40 | 169.7 |
| Chrysemys picta bellii | Painted turtle | XP_008177865.1 | 135 | 35 | 320.5 |
Clinical Significance
A bipolar disorder association study identified C11orf86 as one of many genes found in a region of linkage disequilibrium on chromosome 11. Despite evidence of some association, C11orf86 was not found to be in an area of particular significance.[21] C11orf86 is down-regulated from non-neoplastic mucosa to adenomas and carcinomas,[22] down-regulated in renal cell carcinoma,[23] and harbors chromosomal gains that are significantly associated with pure mucinous subtypes in mucinous carcinoma.[24]
References
- ↑ "C11orf86 chromosome 11 open reading frame 86 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2016-05-09.
- 1 2 "uncharacterized protein C11orf86 [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2016-04-24.
- ↑ "Gene neighbors for C11orf86 - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2016-05-09.
- ↑ "Genomatix Software Suite - Scientific Analysis of genomic data". www.genomatix.de. Retrieved 2016-04-24.
- ↑ "Tissue expression of C11orf86 - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2016-05-09.
- ↑ "EST Profile - Hs.232604". www.ncbi.nlm.nih.gov. Retrieved 2016-04-07.
- ↑ "AceView - Entry for C11orf86". www.ncbi.nlm.nih.gov. Retrieved 2016-05-09.
- ↑ "Homo sapiens chromosome 11 open reading frame 86 (C11orf86), mRNA - Nucleotide - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2016-05-09.
- 1 2 "SAPS". SDSC Biology Workbench. Retrieved 2016-05-08.
- ↑ "PI". SDSC Biology Workbench. Retrieved 2016-05-08.
- ↑ "NCBI CDD Conserved Protein Domain DUF4633". www.ncbi.nlm.nih.gov. Retrieved 2016-02-28.
- ↑ "CDART". www.ncbi.nlm.nih.gov. Retrieved 2016-05-08.
- ↑ "NetPhos 2.0 Server". www.cbs.dtu.dk. Retrieved 2016-05-08.
- ↑ "NetOGlyc 4.0 Server". www.cbs.dtu.dk. Retrieved 2016-05-08.
- ↑ "PELE". SDSC Biology Workbench. Retrieved 2016-04-24.
- ↑ "Ali2D". Bioinformatics Toolkit. Max Planck Institute for Developmental Biology. Retrieved 2016-04-24.
- ↑ "PSORT II Prediction". psort.hgc.jp. Retrieved 2016-05-09.
- ↑ "Motif Scan". myhits.isb-sib.ch. Retrieved 2016-05-08.
- ↑ "BLAST - Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2016-02-28.
- ↑ "TimeTree :: The Timescale of Life". www.timetree.org. Retrieved 2016-02-28.
- ↑ Sklar, Pamela; Ripke, Stephan; Scott, Laura J; Andreassen, Ole A; Cichon, Sven; Craddock, Nick; Edenberg, Howard J; Nurnberger, John I; Rietschel, Marcella. "Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4". Nature Genetics. 43 (10): 977–983. doi:10.1038/ng.943. PMC 3637176
 . PMID 21926972.
. PMID 21926972. - ↑ Kim, Hyunki; Eun, Jung Woo; Lee, Hanna; Nam, Suk Woo; Rhee, Hwanseok; Koh, Kwi Hye; Kim, Hoguen (2011-04-01). "Gene expression changes in patient-matched gastric normal mucosa, adenomas, and carcinomas". Experimental and Molecular Pathology. 90 (2): 201–209. doi:10.1016/j.yexmp.2010.12.004.
- ↑ Hidaka, Hiedo; Seki, Naohiko; Yoshino, Hirofumi; Yamasaki, Takeshi; Yamada, Yasutoshi; Nohata, Nijiro; Fuse, Miki; Nakagawa, Masayuki; Enokida, Hideki (2012-01-01). "Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma". Oncotarget. 3 (1): 44–57. doi:10.18632/oncotarget.417. ISSN 1949-2553. PMC 3292891
 . PMID 22294552.
. PMID 22294552. - ↑ Lacroix-Triki, Magali; Suarez, Paula H; MacKay, Alan; Lambros, Maryou B; Natrajan, Rachael; Savage, Kay; Geyer, Felipe C; Weigelt, Britta; Ashworth, Alan (2010-11-01). "Mucinous carcinoma of the breast is genomically distinct from invasive ductal carcinomas of no special type". The Journal of Pathology. 222 (3): 282–298. doi:10.1002/path.2763. ISSN 1096-9896.