C9orf135
Background
C9orf135 is a gene that is 229 amino acids long and is located on Chromosome 9 within Homo sapiens at exactly 9q12.21.[1] It has a transmembrane domain from amino acids 124-140 and it also has a glycosylation site at amino acid 75. C9orf135 is part of GRCh37 gene on Chromosome 9 and contained within the domain of unknown function superfamily 4572.[2] Also, c9orf135 is known by the name of LOC138255 which is a description of the gene location on Chromosome 9.1.[3]
One particular disease called premature ovarian failure has contained evidence of the c9orf135 gene in affected women.[4] In these women, an autosomal recessive microduplication occurs which may have a link to premature ovarian failure. There has also been a link between Parkinson’s disease in which the c9orf135 gene had a Single Nucleotide Polymorphism (SNP) that appeared to express a statistically significant mutation that was seen on a Manhattan plot.[5] More evidence and research must be done to understand if c9orf135 does in fact relate to Parkinson’s disease.[5]
mRNA

The mRNA of c9orf135 is 906 nucleotides in length.[6] The secondary mRNA structure is composed of both beta-sheets and alpha-helices.[7] The 5' and 3' Untranslated regions (UTR) were looked at for c9orf135. Both of the UTR had hairpin loops that were present.[8][8] The 3' UTR has a total of 123 nucleotides and the 5' UTR has a total of 18 nucleotides.
-
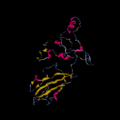
Secondary structure of c9orf135
-

5' UTR loop structure of c9orf135 of mRNA
-

3' UTR loop structure of c9orf135 mRNA
Protein
Properties of c9orf135
c9orf135 is likely a nuclear protein because it has properties that match attributes of nuclear proteins rather than secretory pathways.[9][10] Furthermore, there is a nuclear localization signal PEKVKKL from amino acid 67 to 73 on c9orf135.[11] C9orf135 is souluble with an average hydrophobicity of -0.772. The hydrophobicity is a negative value which explains that it has slightly acidic properties.[12]
Post-translational modifications
Phosphorylation sites were seen at serine for amino acid 7, 50, 86, 98, and 194. Threonine phosphorylation occurred at 34, 129, 155, and 201. Finally, Tyrosine phosphorylation sites occurred at 78, 160, 177, and 209. Also, a N-terminal acetylation site occurred at amino acid 3. Finally, there was a Signal cleavage site between amino acid 11 and 12.[13]
Protein Interaction
PB2 interacts with c9orf135 which was found from a two-hybrid yeast assay. The information provided about PB2 (Polymerase Basic Protein 2) is that it is a viral protein that is involved with the influenza A virus. It is primarily involved in Cap stealing in which it binds the pre-mRNA cap an ultimately cleaves 10-13 nucleotides off. Also, PB2 is important for starting the replication of viral genomes. One final thing PB2 is involved with is the ability to inhibit type 1 interferon. It does this by inhibiting mitochondrial antiviral signaling protein MAVS.[14]
Mutations within c9orf135
Mutations that were present at levels of 0.01 frequency or higher were incorporated into the table. Also, synonymous mutations were not included within the chart. There were 11 different common DNA genome variants of the c9orf135 gene within humans and all of the mutations within those genome variants were compiled into the following chart.[15]
| Location | Amino Acid Position | Mutation | Frequency |
|---|---|---|---|
| 5' UTR | 57 | N/A | 0.386 |
| 5' UTR | 93 | N/A | 0.047 |
| 5' UTR | 138 | N/A | 0.018 |
| Exon | 169 | Missense K30T | 0.018 |
| Exon | 237 | Missense R53K | 0.047 |
| Exon | 456 | Missense E126K | 0.01 |
Gene Expression
c9orf135 is expressed in connective tissue and testicular tissue at high levels.[16] It is likely that the expression of c9orf135 is expressed at low levels throughout human cells. It was also found that c9orf135 is found at significantly higher levels in the adult human umbilical cord versus the fetal human umbilical cord.[17] Furthermore, in women that have an ovarian adenocarcinoma the expression of c9orf135 is much higher in the epithelial cells within the ovaries.[18] Finally, individuals that have polycystic ovarian syndrome have a lower expression of c9orf135 than those people who do not have the condition.[19]
-

Fetal and Adult Umbilical cord expression
-
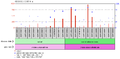
Ovarian normal surface epithelia and ovarian cancer epithelial cells
-
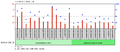
Polycystic ovarian syndrome versus obese women regarding c9orf135.
Amino Acid Quantity
In comparison, Mus musculus (House Mouse) and Pteropus alecto (Black Flying Fox) c9orf135 were also looked at. There were no significant amino acids that differed in c9orf135 from the rest of the mouse body. However, in the Black Flying Fox, it was Valine poor and Tryptophan rich. As seen from the human results, the Black Flying fox only shared the Trytophan surplus results. The House Mouse and Black Flying Fox were both used because they shared 64% and 79% similarity in the c9orf135 genome respectively. Analysis can show that Alanine and Tyrosine could predict points of interest because they both contained results differing from the rest of the human gene averages.[12]
| Amino Acid of Interest | Compositional Percentage |
Compared to normal protein amounts in H. sapiens |
|---|---|---|
| Alanine | 3.1% | Less than Average |
| Tyrosine | 5.2% | More than Average |
| Trytophan | 3.1% | More than average |
Homology
c9orf135 is conserved through eukaryotes, ranging from mammals, reptiles and Annelida.
Orthologs
The orthologs of c9orf135 were sequenced in BLAST and 20 orthologs were picked. The orthologs were all multicellular organisms and were limited to aquatic animals, reptiles, amphibians, and warm-blooded animals. Also, protists, bacteria, archea, and fungi did not have orthologs. However, no paralogs were found when c9orf135 was sequenced in BLAST. Please refer to the spreadsheet for the complete list of orthologs to c9orf135. Time tree was a program that was used to find the evolutionary branching shown in MYA[20] There were no paralogs found for c9orf135.
| Genus/Species | Common Name | Divergence from Humans (MYA) | Accession Number | Amino Acid Length | Sequence Identity | Sequence Similarity |
|---|---|---|---|---|---|---|
| Homo sapiens | Human | -- | Q5VTT2 | 229 | -- | -- |
| Pongo abelii | Sumatran Orangutan | 15.8 | XP_002819904 | 206 | 86% | 87% |
| Rhinopithecus roxellana | Golden Snub-Nosed Monkey | 29.1 | XP_010361250 | 229 | 93% | 95% |
| Mus musculus | House Mouse | 90.9 | EDL41604 | 228 | 64% | 73% |
| Pteropus alecto | Black Flying Fox | 97.5 | XP_785964 | 230 | 79% | 86% |
| Equus przewalskii | Przewalski's horse | 97.5 | XP_008504806 | 183 | 77% | 86% |
| Panthera tigris altaica | Siberian Tiger | 97.5 | XP_007077537 | 187 | 73% | 83% |
| Ovis aries | Sheep | 97.5 | XP_014948670 | 207 | 69% | 77% |
| Elephantulus edwardii | Cape Elephant Shrew | 105 | XP_006894485 | 254 | 72% | 82% |
| Pelodiscus sinensis | Chinese Softshell Turtle | 320.5 | XP_006137902 | 217 | 55% | 68% |
| Gekko japonicus | Gekko | 320.5 | XP_015275999 | 221 | 52% | 64% |
| Alligator mississippiensis | American Alligator | 320.5 | XP_014464144 | 212 | 51% | 64% |
| Ophiophagus hannah | King Cobra | 320.5 | ETE61720 | 215 | 43% | 59% |
| Salmo Salar | Atlantic Salmon | 429.6 | XP_013998840 | 99 | 34% | 55% |
| Esox lucius | Northern Pike | 429.6 | XP_010901691 | 154 | 30% | 47% |
| Branchiostoma floridae | Lancelet | 733 | XP_002591786 | 221 | 45% | 59% |
| Strongylocentrotus purpuratus | Sea Urchin | 747.8 | XP_785964 | 241 | 47% | 62% |
| Saccoglossus kowalevskii | Acorn Worm | 747.8 | XP_002733410 | 153 | 38% | 58% |
| Lingula anatina | Ocean Clam | 847 | XP_013398605 | 220 | 43% | 59% |
| Crassostrea gigas | Pacific Oyster | 847 | XP_011426944 | 215 | 40% | 57% |
| Helobdella robusta | Leech | 847 | XP_009019861 | 256 | 29% | 44% |
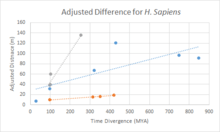
Divergence of c9orf135
In a divergence comparison of c9orf135 with fast diverging cytochrome C, and slow diverging fibrinogen is shown in the chart. Overall, c9orf135 has diverged at a fairly quick pace with it being significantly quicker than fibrinogen and slightly slower than cytochrome C.
References
- ↑ NCBI, Protein, http://www.ncbi.nlm.nih.gov/protein/Q5VTT2.1
- ↑ BLAST protein sequence, c9orf135, http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ↑ Result Filters. (n.d.). Retrieved February 07, 2016, from http://www.ncbi.nlm.nih.gov/protein/Q5VTT2.1
- ↑ Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. McGuire, Megan M; Bowden, Wayne; Engel, Natalie J; Ahn, Hyo Won; Kovanci, Ertug et al.(2011) Fertility and sterility vol.95(5)p.1595-600
- 1 2 Genomic determinants of motor and cognitive outcomes in Parkinson's disease. Chung, Sun Ju; Armasu, Sebastian M; Biernacka, Joanna M; Anderson, Kari J; Lesnick, Timothy G et al.(2012)Parkinsonism & related disorders vol. 18(7)p.881-6
- ↑ NCBI, Nucleotide, http://www.ncbi.nlm.nih.gov/nuccore/809279636?report=fasta
- ↑ I-Tasser, http://zhanglab.ccmb.med.umich.edu/I-TASSER/
- 1 2 Sfold, Srna, http://sfold.wadsworth.org/cgi-bin/showcentroid.pl
- ↑ YLoc http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi?id=86ac5008fd25bd20a065f49a970fd19c
- ↑ SOSUI Classification and Secondary Structure Prediction of Membrane Proteins, http://harrier.nagahama-i-bio.ac.jp/sosui/
- ↑ PSORT II, PSORT II Prediction, http://psort.hgc.jp/cgi-bin/runpsort.pl
- 1 2 SDSC Biology Workbench, SAPS, http://seqtool.sdsc.edu/CGI/BW.cgi#!
- ↑ ExPasy, Sibs bioinformatics analysis, http://www.expasy.org/
- ↑ IntAct http://www.ebi.ac.uk/intact/interactions?conversationContext=2
- ↑ NCBI SNP Geneview http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?geneId=138255&ctg=NT_008470.20&mrna=XM_011518232.1&prot=XP_011516534.1&orien=forward
- ↑ EST profile, NCBI, http://www.ncbi.nlm.nih.gov/nuccore/NM_001308086.1
- ↑ Geo Profile, NCBI http://www.ncbi.nlm.nih.gov/geoprofiles/78193260
- ↑ Geo Profile, Ovarian normal surface epithelia and the ovarian cancer epithelial cells, http://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS3592:236519_at
- ↑ Geo Profile, Obese women with polycystic ovary syndrome and obese, healthy women: skeletal muscle, http://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS4133:243610_at
- ↑ Hedges SB, Marin J, Suleski M, Paymer M & Kumar S (2015) Tree of Life Reveals Clock-Like Speciation and Diversification. Mol Biol Evol 32: 835-845