MicX sRNA
| MicX Vibrio cholerae sRNA | |
|---|---|
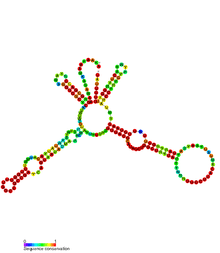 | |
| Secondary structure of MicX sRNA. | |
| Identifiers | |
| Symbol | MicX |
| Rfam | RF01808 |
| Other data | |
| RNA type | sRNA |
| Domain(s) | Vibrio cholerae |
MicX sRNA (formerly known as A10)[1] is a small non-coding RNA found in Vibrio cholerae.[2] It was given the name MicX as it has a similar function to MicA, MicC and MicF in E. coli.[3] MicX sRNA negatively regulates an outer membrane protein (coded for by VC0972) and also a component of an ABC transporter (gene VC0620).[4] These interactions were predicted and then confirmed using a DNA microarray.[2]
MicX was identified through a bioinformatics screen of V. cholerae having been previously predicted.[1] Levels of transcription of this sRNA were compared under several conditions: it was found to be expressed on all tested mediums; richer mediums slightly reduced transcription; repression of certain sigma factors (δS and δE) did not change transcription but it was dramatically reduced in the absence of Hfq protein.[2] This observation is in accordance with other sRNA expression patterns.[5][6]
The MicX RNA gene overlaps with VCA0943 - a gene coding for a maltose transporter permease - but the ribonuclease RNAase E processes the MicX transcript to create an active and stable form containing only the VCA0943 3' UTR.[2]
See also
References
- 1 2 Livny, Jonathan; Fogel, Michael A.; Davis, Brigid M.; Waldor, Matthew K. (2005). "sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes". Nucleic Acids Research. 33 (13): 4096–105. doi:10.1093/nar/gki715. PMC 1180744
 . PMID 16049021.
. PMID 16049021. - 1 2 3 4 Davis, Brigid M.; Waldor, Matthew K. (2007). "RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA". Molecular Microbiology. 65 (2): 373–85. doi:10.1111/j.1365-2958.2007.05796.x. PMC 1976385
 . PMID 17590231.
. PMID 17590231. - ↑ Mizuno, T.; Chou, M. Y.; Inouye, M. (1984). "A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA)". Proceedings of the National Academy of Sciences. 81 (7): 1966–70. doi:10.1073/pnas.81.7.1966. PMC 345417
 . PMID 6201848.
. PMID 6201848. - ↑ Song, Tianyan; Mika, Franziska; Lindmark, Barbro; Liu, Zhi; Schild, Stefan; Bishop, Anne; Zhu, Jun; Camilli, Andrew; et al. (2008). "A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles". Molecular Microbiology. 70 (1): 100–11. doi:10.1111/j.1365-2958.2008.06392.x. PMC 2628432
 . PMID 18681937.
. PMID 18681937. - ↑ Sledjeski, Darren D.; Whitman, Christina; Zhang, Aixia (2001). "Hfq Is Necessary for Regulation by the Untranslated RNA DsrA". Journal of Bacteriology. 183 (6): 1997–2005. doi:10.1128/JB.183.6.1997-2005.2001. PMC 95095
 . PMID 11222598.
. PMID 11222598. - ↑ Massé, Eric; Escorcia, Freddy E.; Gottesman, Susan (2003). "Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli". Genes & Development. 17 (19): 2374–83. doi:10.1101/gad.1127103. PMC 218075
 . PMID 12975324.
. PMID 12975324.
Further reading
- Gottesman, Susan (2004). "The small RNA regulators of Escherichia coli: roles and mechanisms". Annual Review of Microbiology. 58: 303–28. doi:10.1146/annurev.micro.58.030603.123841. PMID 15487940.
- Schlüter, Jan-Philip; Reinkensmeier, Jan; Daschkey, Svenja; Evguenieva-Hackenberg, Elena; Janssen, Stefan; Jänicke, Sebastian; Becker, Jörg D.; Giegerich, Robert; Becker, Anke (2010). "A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti". BMC Genomics. 11: 245. doi:10.1186/1471-2164-11-245. PMC 2873474
 . PMID 20398411.
. PMID 20398411. - Sittka, Alexandra; Sharma, Cynthia M.; Rolle, Katarzyna; Vogel, Jörg (July 2009). "Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes". RNA Biology. 6 (3): 266–75. doi:10.4161/rna.6.3.8332. PMID 19333007.