Proteorhodopsin
| Proteorhodopsin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Proteorhodopsin Cartoon Visualization by ELViture | |||||||||
| Identifiers | |||||||||
| Symbol | Bac_rhodopsin | ||||||||
| Pfam | PF01036 | ||||||||
| InterPro | TCDB | ||||||||
| PROSITE | PDOC00291 | ||||||||
| SCOP | 2brd | ||||||||
| SUPERFAMILY | 2brd | ||||||||
| TCDB | 3.E.1 | ||||||||
| OPM superfamily | 6 | ||||||||
| OPM protein | 4hyj | ||||||||
| |||||||||
Proteorhodopsin (also known as pRhodopsin) is a family of over 50 photoactive retinylidene proteins, a larger family of transmembrane proteins that use retinal as a chromophore for light-mediated functionality, in this case, a proton pump. Some homologues exist as pentamers or hexamers. pRhodopsin is found in marine planktonic bacteria, archaea and eukaryotes (protae), but was first discovered in bacteria.[1][2][3][4]
Its name is derived from the Ancient Greek Πρωτεύς (Proteus), an early sea god mentioned by Homer as "Old Man of the Sea", Ῥόδος (rhódon) for "rose", due to its pinkish color, and ὄψις (opsis) for "sight". Some members of the family, Homologous rhodopsin-like pigments, i.e. bacteriorhodopsin (of which there are more than 800 types) have Sensory Functions like opsins, integral for visual phototransduction. Many of these sensory functions are unknown – for example, the function of Neuropsin in the human retina.[5] Members are known to have different absorption spectra including green and blue visible light.[6][7][8][9][10][11]
Discovery
Proteorhodopsin (PR or pRhodopsin) was first discovered in 2000 within a bacterial artificial chromosome from previously uncultivated marine γ-proteobacteria, still only referred to by their ribotype metagenomic data, SAR86. The research was a cooperative effort between four parties: Oded Beja, Marcelino T. Suzuki, and Edward F. DeLong at Monterey Bay Aquarium Research Institute (Mosslanding, CA), L. Aravind and Eugene V Koonin at the National Center for Biotechnology Information (Bethesda, MD), Andrew Hadd, Linh P. Nguyen, Stevan B. Jovanovich, Christian M. Gates, and Rober A Feldman at Molecular Dynamics (Sunnyvale, CA), and finally John and Elena Spudich at the Department of Microbiology and Molecular Genetics at the University of Texas Medical School. More species of γ-proteobacteria, both Gram positive and negative, were found to express the protein.[1]
Distribution
Samples of proteorhodopsin expressing bacteria have been obtained from the Eastern Pacific Ocean, Central North Pacific Ocean and Southern Ocean, Antarctica.[12] Subsequently, genes of proteorhodopsin variants have been identified in samples from the Mediterranean, Red Seas, the Sargasso Sea, and Sea of Japan, and the North Sea.[4][6]
Proteorhodopsin variants are not spread randomly, but disperse along depth gradients based on the maximal absorption-tuning of the particular holoprotein sequence; this is mainly due to the electromagnetic absorption by water which creates wavelength gradients relative to depth. Oxyrrhis marina is a Dinoflagellate protist with green-absorbing proteorhodopsin (a result of the L109 Group) that exists mostly in shallow tide pools and shores, where green light is still available. Karlodinium micrum, another dinolagelate, expresses a blue tuned proteorhodopsin (E109) which may be related to its deep water vertical migrations.[3] O.Marina was originally believed to be a heterotroph, however the proteorhodopsin may well partake in a functionally significant manner, as it was the most abundantly expressed nuclear gene and, furthermore, is dispersed unevenly in the organism, suggesting some organelle membrane function. Previously the only eukaryotic solar energy transducing proteins were Photosystem I and Photosystem II. It has been hypothesized that lateral gene transfer is the method by which proteorhodopsin has made its way into numerous phyla. Bacteria, archea and eukarya all colonize the photic zone where they come to light; Proteorhodopsin has been able to disseminate through this zone, but not to other portions of the water column.[3][4][9][13][14]
Taxonomy
Proteorhodopsin belongs to a family of similar retinylidene proteins, most similar to its archeal homologes halorhodopsin and bacteriorhodopsin. Sensory Rhodopsin was discovered by Franz Christian Boll in 1876.[11][15] Bacteriorhodopsin was discovered in 1971 and named in 1973 and is currently only known to exist in the archea domain, not bacteria.[16] Halorhodopsin was first discovered and named in 1977. [17] Bacteriorhodopsin and Halorhodopsin both only exist in the Archea domain whereas proteorhodopsin spans bacteria, archea, and eukaryotes. Proteorhodopsin shares seven transmembrane α-helicies retinal covalently linked by a Schiff base mechanism to a lysine residue in the seventh helix (helix G). Bacteriorhodopsin, like proteorhodopsin, is a light-driven proton pump. Sensory Rhodopsin is a G-coupled protein involved in sight.[1][17]
Active site

In comparison with its better-known archaeal homolog bacteriorhodopsin, most of the active site residues of known importance to the bacteriorhodopsin mechanism are conserved in proteorhodopsin. Sequence similarity is not significantly conserved however, from either halo- or bacterio- rhodopsin. Homologues of the active site residues Arg82, Asp85 (the primary proton acceptor), Asp212 and Lys216 (the retinal Schiff base binding site) in bacteriorhodopsin are conserved as Arg94, Asp97, Asp227 and Lys231 in proteorhodopsin. However, in proteorhodopsin, there are no carboxylic acid residues directly homologous to Glu194 or Glu204 of bacteriorhodopsin (or Glu 108 and 204 depending on the bacRhodopsin variant), which are thought to be involved in the proton release pathway at the extracellular surface. However, Asp97 and Arg94 may replace this functionality without the close residue proximity as in bacteriorhodopsin. The department of chemistry at Syracuse University decisevely showed Asp97 cannot be the proton release group as the release happened at forcing conditions under which the aspartic acid group remained protonated.[18][19][20][21]
Ligand
The Rhodopsin haloprotein family shares the ligand Retinal, Vitamin A Aldehyde, one of the many types of Vitamin A. Retinal is a conjugated poly-unsaturatedchromophore (polyene), obtained from carnivorous diet or by the carotene pathway (β-carotene 15,15'-monoxygenase).
Function
Proteorhodopsin functions throughout the Earth's oceans as a light-driven H+ pump, by a mechanism similar to that of bacteriorhodopsin. As in bacteriorhodopsin, the retinal chromophore of proteorhodopsin is covalently bound to the apoprotein via a protonated Schiff base at Lys231. The configuration of the retinal chromophore in unphotolyzed proteorhodopsin is predominantly all-trans[18] , and isomerizes to 13-cis upon illumination with light. Several models of the complete proteorhodopsin photocycle have been proposed, based on FTIR and UV–visible spectroscopy; they resemble established photocycle models for bacteriorhodopsin.[18][20][21][22] Complete proteorhodopsin based photosystems have been discovered and expressed in E. coli, giving them additional light mediated energy gradient capability for ATP generation without external need for retinal or precursors; with the PR, gene five other proteins code for the photopigment biosynthetic pathyway.[23]
Genetic engineering
If the gene for proteorhodopsin is inserted into E. coli and retinal is given to these modified bacteria, then they will incorporate the pigment into their cell membrane and will pump H+ in the presence of light. A deep purple is representative of clearly transformed colonies, due to light absorption. Proton gradients can be used to power other membrane protein structures or used to acidify a vesicle type organelle.[1] It was further demonstrated that the proton gradient generated by proteorhodopsin could be used to generate ATP.[23]
Gallery
-
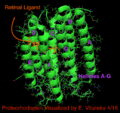
Holoenzyme (Green) with helicies A-G labeled (purple) as well as Retinal ligand (orange)
-

Surface visualization of Proteorhodopsin showing terminals
-

Visualization of the retinal bound active site of the 2L6X protein structure of pRhodopsin, residues color coded and labeled by activity, ligand is orange.
-

2L6x In-Active-Site Cartoon Color Coded and Labeled Visualization, D and E Helicies hidden for vantage, Retinal ligand binding site
References
- 1 2 3 4 Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF (Sep 2000). "Bacterial rhodopsin: evidence for a new type of phototrophy in the sea". Science. 289 (5486): 1902–6. Bibcode:2000Sci...289.1902B. doi:10.1126/science.289.5486.1902. PMID 10988064.
- ↑ Lin S, Zhang H, Zhuang Y, Tran B, Gill J (Nov 2010). "Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates". Proceedings of the National Academy of Sciences of the United States of America. 107 (46): 20033–8. Bibcode:2010PNAS..10720033L. doi:10.1073/pnas.1007246107. PMID 21041634.
- 1 2 3 Slamovits CH, Okamoto N, Burri L, James ER, Keeling PJ (2011). "A bacterial proteorhodopsin proton pump in marine eukaryotes". Nature Communications. 2 (2): 183. Bibcode:2011NatCo...2E.183S. doi:10.1038/ncomms1188. PMID 21304512.
- 1 2 3 Frigaard NU, Martinez A, Mincer TJ, DeLong EF (Feb 2006). "Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea". Nature. 439 (7078): 847–50. Bibcode:2006Natur.439..847F. doi:10.1038/nature04435. PMID 16482157.
- ↑ Buhr ED, Yue WW, Ren X, Jiang Z, Liao HW, Mei X, Vemaraju S, Nguyen MT, Reed RR, Lang RA, Yau KW, Van Gelder RN (20 Oct 2015). ""Neuropsin (OPN6)-mediated photoentrainment of local circadian oscilators in mammalian retina and cornea".". Proceedings of the National Academy of Sciences. 112 (42): 13093–13098. doi:10.1073/pnas.1516259112. PMC 4620855
 . PMID 26392540.
. PMID 26392540. - 1 2 Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF (Jun 2001). "Proteorhodopsin phototrophy in the ocean". Nature. 411 (6839): 786–9. doi:10.1038/35081051. PMID 11459054.
- ↑ Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Béjà O (Apr 2003). "Diversification and spectral tuning in marine proteorhodopsins". The EMBO Journal. 22 (8): 1725–31. doi:10.1093/emboj/cdg183. PMC 154475
 . PMID 12682005.
. PMID 12682005. - ↑ Kelemen BR, Du M, Jensen RB (Dec 2003). "Proteorhodopsin in living color: diversity of spectral properties within living bacterial cells". Biochimica et Biophysica Acta. 1618 (1): 25–32. doi:10.1016/j.bbamem.2003.10.002. PMID 14643930.
- 1 2 Sabehi G, Kirkup BC, Rozenberg M, Stambler N, Polz MF, Béjà O (May 2007). "Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas". The ISME Journal. 1 (1): 48–55. doi:10.1038/ismej.2007.10. PMID 18043613.
- ↑ Encyclopedia of the Neruological Sciences. Academic Press. 29 April 2014. p. 441. ISBN 978-0-12-385158-1.
- 1 2 Giese, Arthur C (Sep 2013). Photophysiology: General Principles; Action of Light on Plants. Elsevier. p. 9. ISBN 978-1-4832-6227-7.
- ↑ Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (Apr 2004). "Environmental genome shotgun sequencing of the Sargasso Sea". Science. 304 (5667): 66–74. Bibcode:2004Sci...304...66V. doi:10.1126/science.1093857. PMID 15001713.
- ↑ Giovannoni, SJ; Bibbs, L; Cho, JC; Stapels, MD; Desiderio, R; Vergin, KL; Rappé, MS; Laney, S; Wilhelm, LJ; Tripp, HJ; Mathur, EJ; Barofsky, DF (3 November 2005). "Proteorhodopsin in the ubiquitous marine bacterium SAR11.". Nature. 438 (7064): 82–5. doi:10.1038/nature04032. PMID 16267553.
- ↑ Kushwaha, SC; Kates, M (23 August 1973). "Isolation and identification of "bacteriorhodopsin" and minor C40-carotenoids in Halobacterium cutirubrum.". Biochimica et Biophysica Acta. 316 (2): 235–43. doi:10.1016/0005-2760(73)90013-1. PMID 4741911.
- ↑ Encyclopedia of the Neurological Sciences. Academic Press. Apr 2014. p. 441. ISBN 978-0-12-385158-1.
- ↑ Oesterhelt, D; Stoeckenius, W (29 September 1971). "Rhodopsin-like protein from the purple membrane of Halobacterium halobium.". Nature: New biology. 233 (39): 149–52. doi:10.1038/newbio233149a0. PMID 4940442.
- 1 2 Matsuno-Yagi, A; Mukohata, Y (9 September 1977). "Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation.". Biochemical and Biophysical Research Communications. 78 (1): 237–43. doi:10.1016/0006-291x(77)91245-1. PMID 20882.
- 1 2 3 Dioumaev AK, Brown LS, Shih J, Spudich EN, Spudich JL, Lanyi JK (Apr 2002). "Proton transfers in the photochemical reaction cycle of proteorhodopsin". Biochemistry. 41 (17): 5348–58. doi:10.1021/bi025563x. PMID 11969395.
- ↑ Partha R, Krebs R, Caterino TL, Braiman MS (Jun 2005). "Weakened coupling of conserved arginine to the proteorhodopsin chromophore and its counterion implies structural differences from bacteriorhodopsin". Biochimica et Biophysica Acta. 1708 (1): 6–12. doi:10.1016/j.bbabio.2004.12.009. PMID 15949979.
- 1 2 Dioumaev AK, Wang JM, Bálint Z, Váró G, Lanyi JK (Jun 2003). "Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic". Biochemistry. 42 (21): 6582–7. doi:10.1021/bi034253r. PMID 12767242.
- 1 2 Krebs RA, Alexiev U, Partha R, DeVita AM, Braiman MS (Apr 2002). "Detection of fast light-activated H+ release and M intermediate formation from proteorhodopsin". BMC Physiology. 2: 5. doi:10.1186/1472-6793-2-5. PMC 103662
 . PMID 11943070.
. PMID 11943070. - ↑ Xiao Y, Partha R, Krebs R, Braiman M (Jan 2005). "Time-resolved FTIR spectroscopy of the photointermediates involved in fast transient H+ release by proteorhodopsin". The Journal of Physical Chemistry B. 109 (1): 634–41. doi:10.1021/jp046314g. PMID 16851056.
- 1 2 Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF (2007). "Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host". PNAS. 104 (13): 5590–5595. Bibcode:2007PNAS..104.5590M. doi:10.1073/pnas.0611470104. PMC 1838496
 . PMID 17372221.
. PMID 17372221.