U12 minor spliceosomal RNA
| U12 minor spliceosomal RNA | |
|---|---|
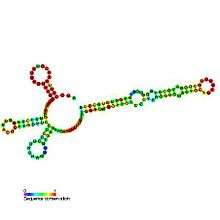 Predicted secondary structure and sequence conservation of U12 | |
| Identifiers | |
| Symbol | U12 |
| Rfam | RF00007 |
| Other data | |
| RNA type | Gene; snRNA; splicing |
| Domain(s) | Eukaryota |
| GO | 0000371 0045131 0005693 |
| SO | 0000399 |
U12 minor spliceosomal RNA is formed from U12 small nuclear (snRNA), together with U4atac/U6atac, U5, and U11 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns. Although the U12 sequence is very divergent from that of U2, the two are functionally analogous.[1] The predicted secondary structure of U12 RNA is published,[2] but the alternative single hairpin in the 3' end shown here seems to better match the alignment of divergent Drosophila melanogaster and Arabidopsis thaliana sequences.[3] The sequences U12 introns that are spliced out are collected in a biological database called the U12 intron database.
References
- ↑ Tarn, WY; Steitz JA (1997). "Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge". Trends Biochem Sci. 22 (4): 132–137. doi:10.1016/S0968-0004(97)01018-9. PMID 9149533.
- ↑ Shukla, GC; Padgett RA (1999). "Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants". RNA. 5 (4): 525–538. doi:10.1017/S1355838299982213. PMC 1369779
 . PMID 10199569.
. PMID 10199569. - ↑ Otake, LR; Scamborova P; Hashimoto C; Steitz JA (2002). "The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila". Mol Cell. 9 (2): 439–446. doi:10.1016/S1097-2765(02)00441-0. PMID 11864616.
External links
This article is issued from Wikipedia - version of the 7/25/2014. The text is available under the Creative Commons Attribution/Share Alike but additional terms may apply for the media files.